Hybrid assembly of ultra-long Nanopore reads augmented with 10x-Genomics contigs: Demonstrated with a human genome - ScienceDirect

SciELO - Brasil - Approaches for <i>in silico</i> finishing of microbial genome sequences Approaches for <i>in silico</i> finishing of microbial genome sequences
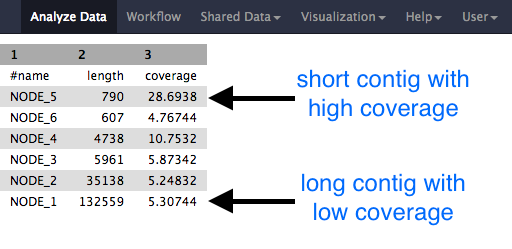
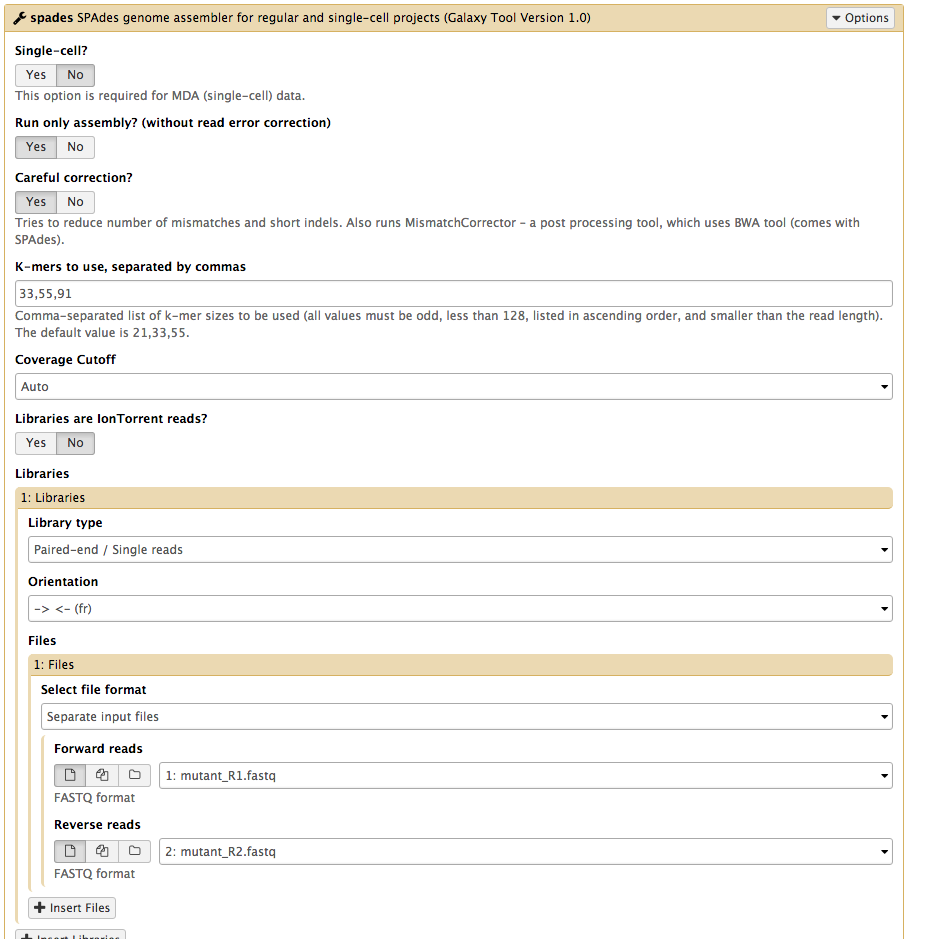
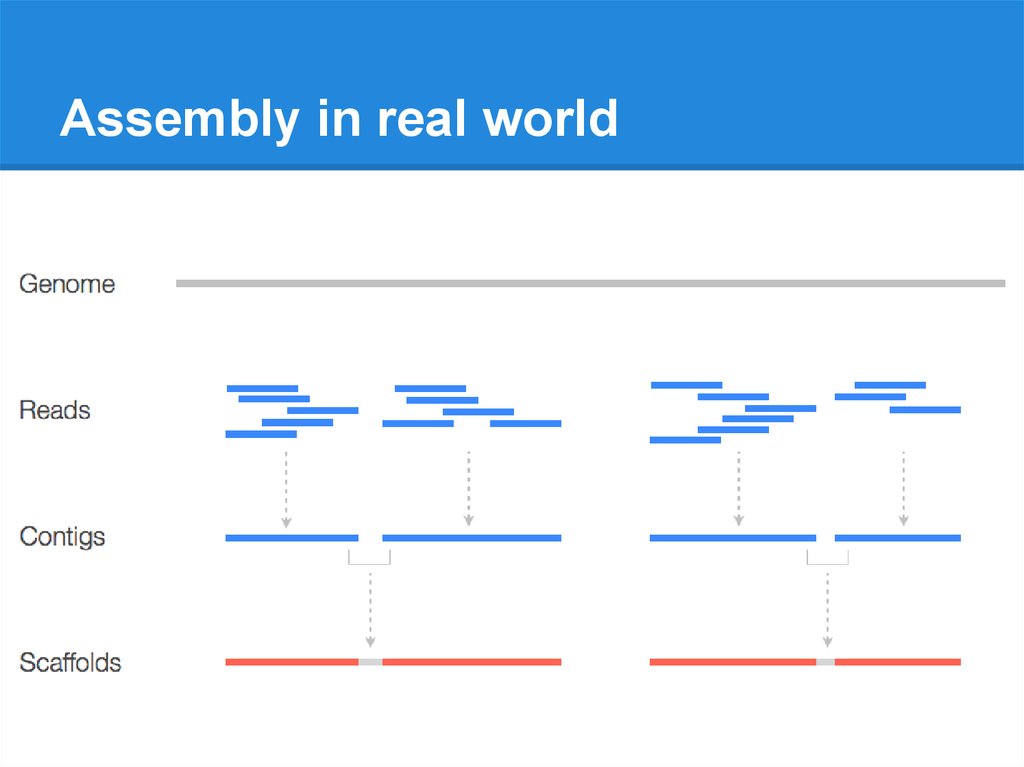
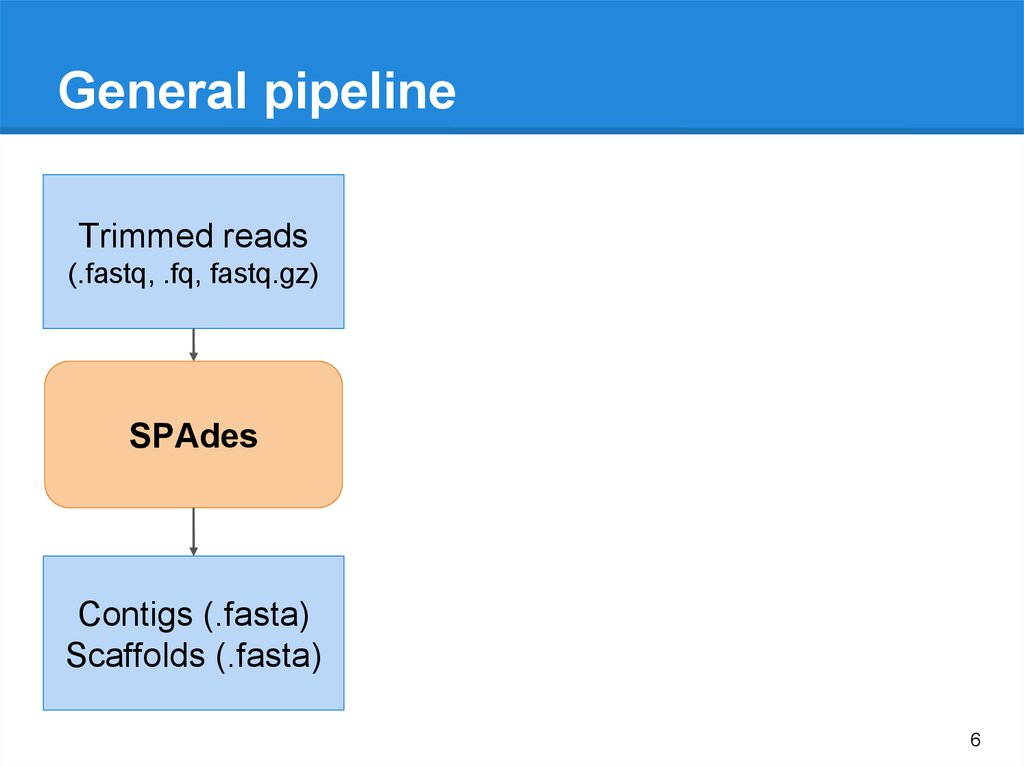
Which one contigs.fasta or scaffolds.fasta should I use for next analysis · Issue #226 · ablab/spades · GitHub

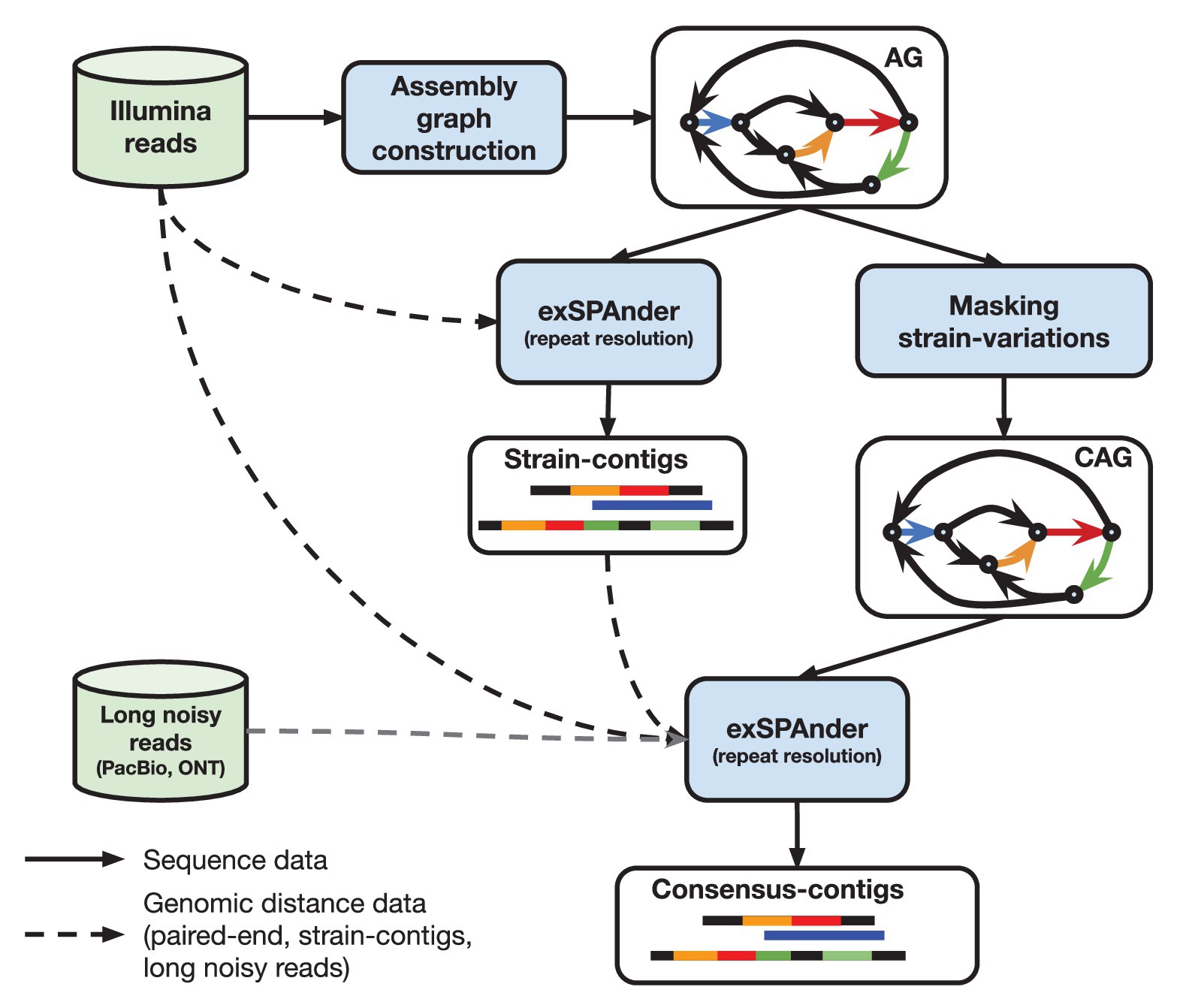
Scaffolding and completing genome assemblies in real-time with nanopore sequencing | Nature Communications

MetaSort untangles metagenome assembly by reducing microbial community complexity | Nature Communications

Reference-based contig ordering. (a) The program takes a set of contigs... | Download Scientific Diagram
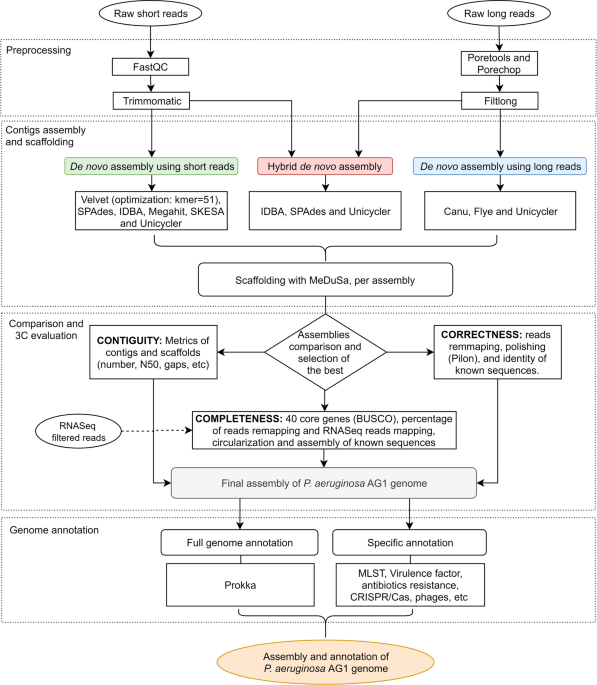
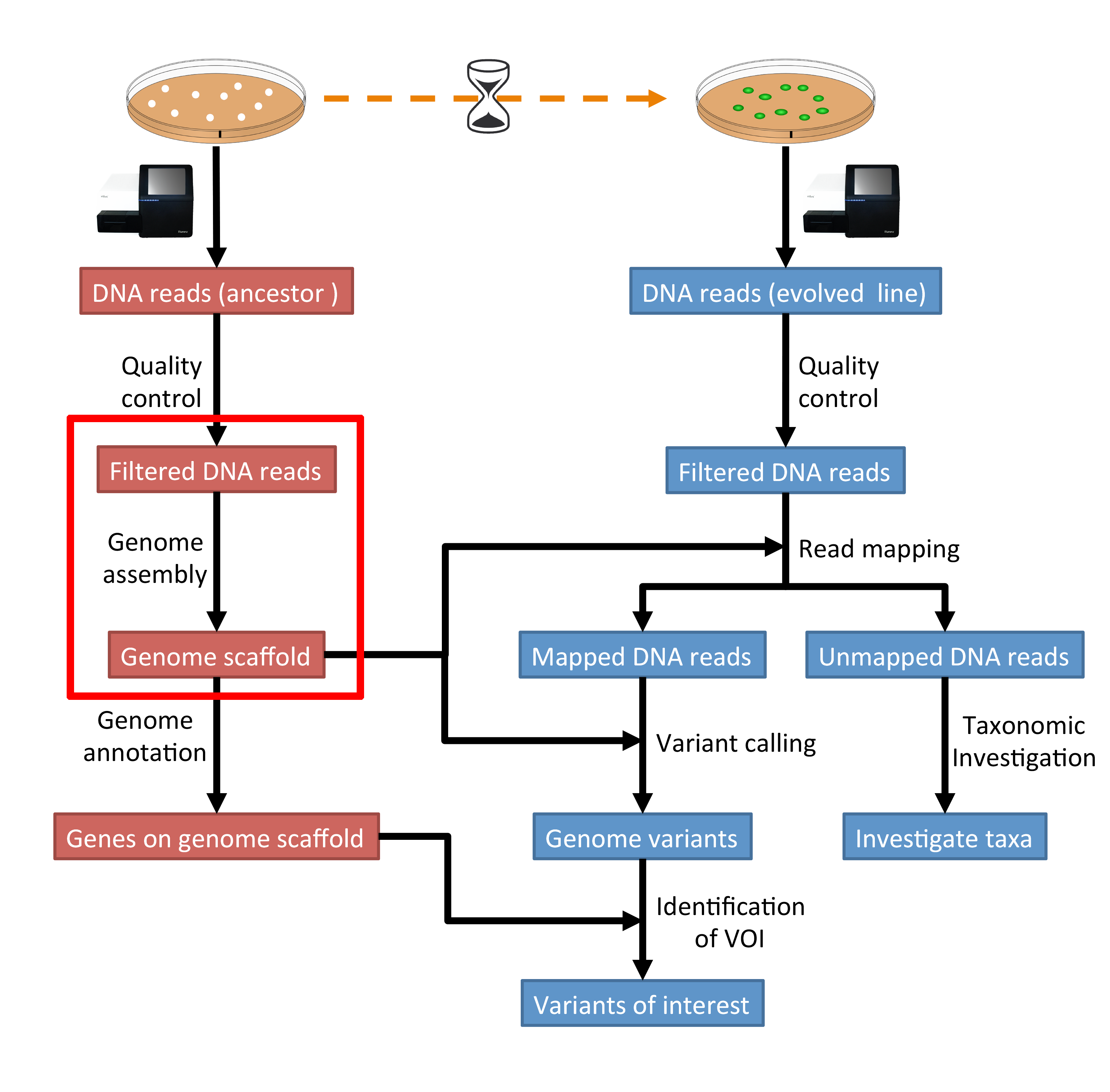
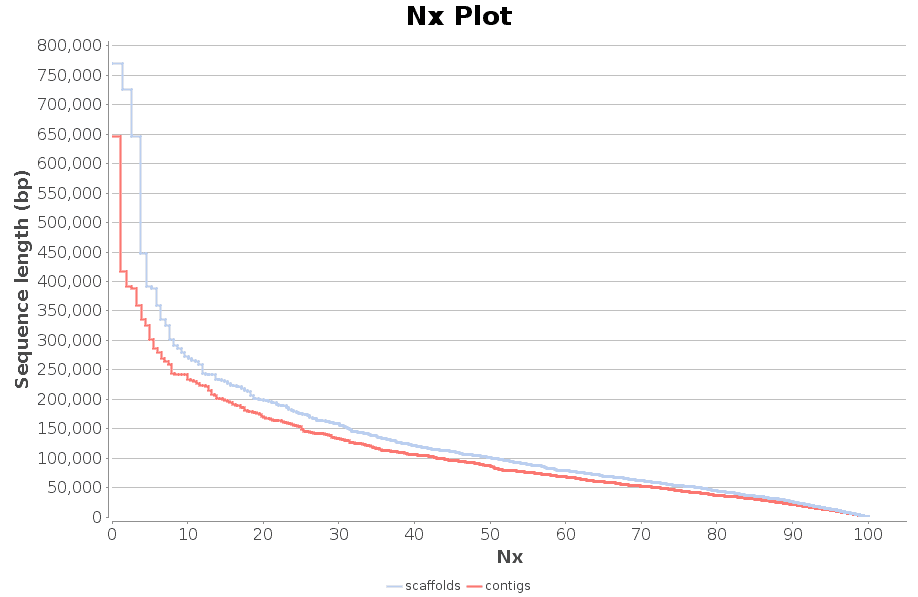
High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers | Scientific Reports